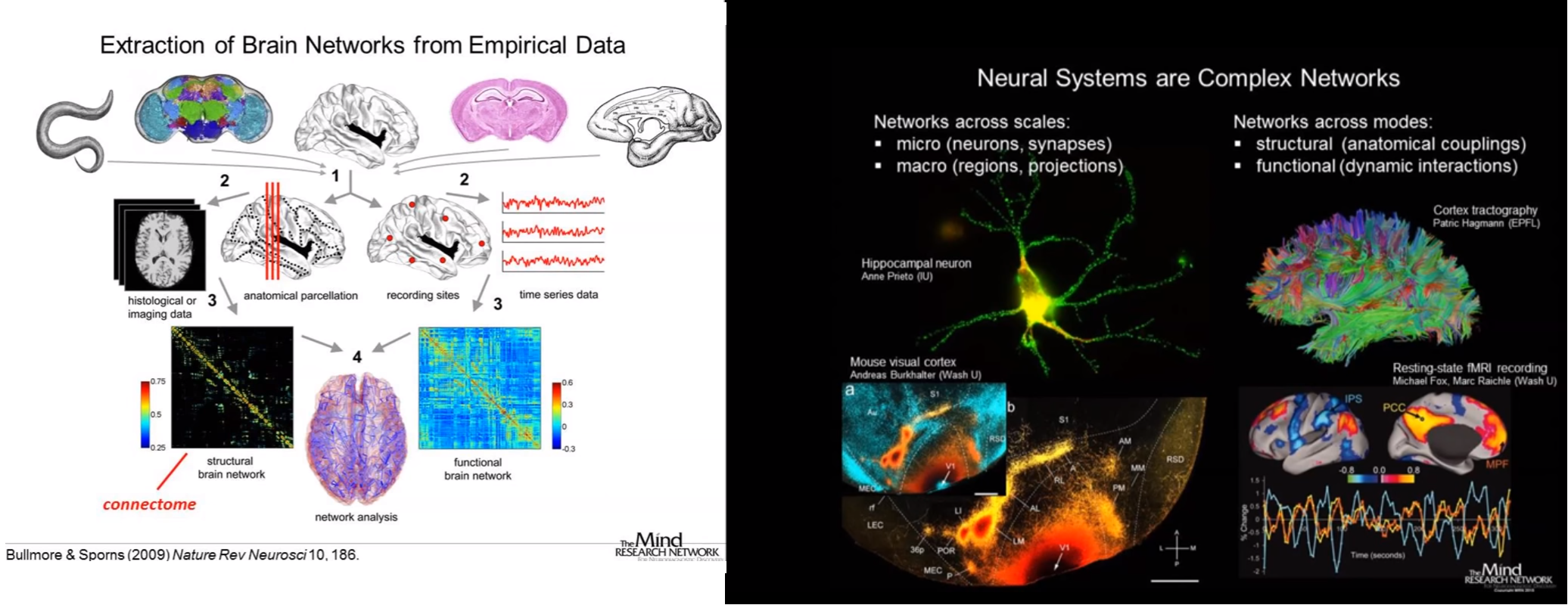


Introduction: The importance of networks has become of central interest in the natural sciences, particularly in the study of complex biological systems including the brain. Modern network approaches are beginning to reveal fundamental principles of brain architecture and function. Connectivity plays an important role in neuroanatomy, neurodevelopment, electrophysiology, functional brain imaging, neural basis of cognition. The analysis of network architecture and connectivity illuminates various problems related to integrative brain functions, e.g., the interaction of neurons in complex physiological responses, the integration of sensory features within and across modalities in cerebral cortex, the changes in sensory input or cognitive tasks, effects of brain trauma or disease, variations in cognitive performance. This new pilot advanced undergraduate-/graduate- level course is an introductory course that will aim to highlight some of the emerging points of contact between neuroscience and network theory and analysis. To understand the structure, behaviour and evolution of a complex system, such as brain, we require not only knowledge of elementary system components but also knowledge of the ways in which these components interact and the emergent properties of their interactions. The increasing availability of large datasets, powerful computers, and statistical analysis methods make it easier than before to record, analyze, and model the behaviour of such systems. Neocortex is a six-layered, folded, sheet-like structure that consists of billions of heavily interconnected neurons. It is the interlaced activity patterns across the neocortical populations of neurons which bestow the capacity to interact intelligently with the environment. Over time much has been learned about the computational properties of single neurons. However, we remain far from understanding how networks of cortical cells coordinate and interact with each other in order to process information and to allow cortical circuits to be modified during learning. Prof. Stelios Smirnakis (Brigham Women's Hospital and Department of Neurology, Harvard Medical School) and Prof. Maria Papadopouli (Department of Computer Science, University of Crete) will discuss the functional sub-network architecture of the neocortex. To better illustrate the main issues, the course will focus on the primary visual cortex of the mouse. There are a lot of experimental data collected from the primary visual cortex of the mouse. Dr. Anna Palagina (Harvard Medical School) collected GCaMP6- and OGB -based datasets using 2-photon imaging from the V1 area (layers 3/4) from mice. These datasets will be employed to present some important concepts and they will be used for analysis in several assignments and projects. The course's aim is to introduce students to the basic biology of the mouse neocortex and it will present graph-theoretical and statistical analysis tools for analyzing experimental data. It will demonstrate different types of networks and their properties.
Course Description The course's aim is to introduce students to the basic biology of the neocortex and present its functional network architecture. To better illustrate the main issues, the course will focus on the primary visual cortex of the mouse. In the first part of the course, it will overview the principles of brain organization, neurophysiology and biophysics of excitable cells, synaptic transmission, network anatomy and physiology and canonical circuits in mouse neocortex. It will then focus on the multi-neuronal computations. The second part will present graph-theoretical and statistical analysis tools for analyzing functional networks. In the third part of the course, experimental methods for probing circuit function using 2 photo imaging, optogenetics, patch clamping in vivo, in vitro will be discussed. An important part of the course will be the project identification and implementation. During the last week, the students will present their project.
Tentative Syllabus| Short Description |
| Introduction of the course Basic concepts on neurons Introductory session for the CS students without a background in neuroscience/biology Slides Prof Maria Papadopouli |
| Statistical Analysis: Correlations, Null Hypothesis Testing Statistics: Hypothesis testing, probabilities, CDFs Temporal correlation measures: Pearson correlation, autocorrelation, periodograms, STTC Slides Modeling Tools, Null Hypothesis, Temporal Correlation Modeling Temporal Correlation, STTC, Kolmogorov Smirnov test Prof Maria Papadopouli |
| Graph-theoretical methods of analysis Graph theory, Graph-theoretical properties: path length, degree of connectivity, diameter, clustering coefficient Network architectures: lattice/regular graphs, random graphs, small world, scale-free networks Slides Graph Theory Prof Maria Papadopouli |
| Fundamentals in neuroscience I Cellular mechanisms of Memory; Monday, October 14 at 11.00a.m. Please note the change in time, only for that day Slides Memory Mechanisms Prof Kiki Sidiropoulou Transmission between neurons; the role of neurotransmitters; basics in cognitive neuroscience Thursday or Friday, October 17/18 (during the lab session) Slides Structure & function of the cerebral cortex (updated, 2021) Dr Manolis Froudarakis (guest lecture) Functional organization of the cortex: from functional columns to cell assemblies Slides Functional-organization of the cortex Dr Vassilis Kehayas |
| Fundamentals in neuroscience II Principles of brain organization, structures of neurons, Neurophysiology and biophysics of excitable cells; synaptic transmission; Network Anatomy, physiology and canonical circuits in mouse neocortex Focus on visual cortex of the mouse - layers - description Functional vs. Anatomical vs. Effective connectivity Slides Early Visual System Biological_Networks, monitoring methods, 2-photon_imaging Prof Stelios Smirnakis |
| Multi-neuronal computations Desirable Computational properties of biological circuits Sensory representations and shannon information theory (To be decided) Prof Stelios Smirnakis |
| Statistical Analysis: Correlations, Null Hypothesis Testing, Regression and Clustering Statistics: Hypothesis testing, probabilities, CDFs Temporal correlation measures: Pearson correlation, autocorrelation, periodograms, STTC Slides Modeling Tools, Null Hypothesis, Temporal Correlation, Temporal Correlation (updated) Modeling Temporal Correlation, STTC, Kolmogorov Smirnov test K-means-Linear Regression-Lasso-Ridge Prof Maria Papadopouli |
| Information Capacity and Learning in Neuroscience Slides Information Capacity and Learning in Neuroscience Prof Ioannis Smirnakis |
| Neural dynamics and learning in Spiking Neural Networks Slides Neural dynamics and learning in Spiking Neural Networks Dr. Angeliki Pantazi (IBM Zurich) |
| Group meetings - Team Work Identification and start of the implementation of the project from 1) literature review, or 2) a small data analysis laboratory project on two photon data that will be provided. Such projects could involve the graph-theoretical analysis and pattern discovery of neuronal circuits in absence epilepsy, under spontaneous conditions, etc Team work under the supervision of Stelios Smirnakis and Maria Papadopouli |
| Life in Networks Presentation URL Life in Networks Professor Christos Papadimitriou (Columbia University) Also, part of the Distinguished Lecture Series, Department of Computer Science, University of Crete |
| Reccurent Quantification Analysis (RQA) Slides Recurrence Quantification Analysis Dr. George Tzagkarakis (guest lecture) |
| Machine Learning Machine-learning algorithms basics, feature extraction, SVD Deep learning Relation of biological networks to computational networks Slides Artificial Neural Networks - Convolutional Neural Networks - Recurrent Neural Networks Dr. Greg Tsagkatakis Generative Adversarial Networks (GANs) Slides Dr Yannis Pantazis (guest lecture) Machine Learning for Graphs - Kernels and node embeddings Prof. Michalis Vazirgiannis |
| 1. Large-scale, spatio-temporal patterning in motor cortex and its role in movement initiation 2. Developing brain-machine interfaces in monkeys and humans Prof Nicholas Hatsopoulos (University of Chicago) |
| Group meetings - Team Work - Implementation & discussions of the project Team work under the supervision of Stelios Smirnakis and Maria Papadopouli |
| Experimental methods for probing circuit function 2 photon imaging; optogenetics etc; patch clamping in vivo, in vitro Learning in neuronal networks Hebbian plasticity Prof Stelios Smirnakis |
|
Douglas Martin - Requirements of circuit code Jiang paper circuit connectivity Ensembles ala Yuste - future questions/ experimental approaches - bottom up vs top-down - how to best decipher circuit function Neuroscience vs. Deep Learning Slides Cortical Circuits Prof Stelios Smirnakis |
| Complex Systems Slides Complex Systems |
| Advanced Computational Topics 1. Discussion of the fundamental paper on a Mathematical Theory of Communication by Shannon 2. The Recurrence Quantification Analysis (RQA) for identifying the different states of a dynamic complex system. a. Marwan, N. (2008). "A Historical Review of Recurrence Plots" . European Physical Journal ST. 164 (1): 3–12. arXiv, Bibcode, DOI b. Marwan, N., Romano, M. C. ,Thiel, M. ,Kurths, J. (2007). "Recurrence Plots for the Analysis of Complex Systems". Physics Reports. 438 (5–6): 237–329. Bibcode, DOI c. Marwan, N., Wessel, N., Meyerfeldt, U., Schirdewan, A., Kurths, J. (2002). "Recurrence Plot Based Measures of Complexity and its Application to Heart Rate Variability Data". Physical Review E. 66 (2): 026702. arXiv, Bibcode, DOI Prof Maria Papadopouli |
| Reconvene for project presentation Workshop 2017 |
| Date | Material |
| - | Probability Theory Basics (Maria Markaki) |
| - | Matlab Basics (Maria Markaki) |
| - | Temporal Correlation STTC (Andreas Sapoutzis) |
| - | Graph Theory Assignment, ReadMe_Graph, Plot_Graph_Matlab Code |
| - | Linear Regression, Linear Regression Examples_Matlab Code & clustering |
| - | SVM (Manthos Kampourakis) |
| - | Neural Networks, Decision Trees, Random Forests |
| - | Biological Networks Literature Review and Discussion |
| - | Principal Component Analysis |
Grading Formula
MAX (10% assignments + 25% final exam + 50% project + 10% report/meetings + 5% workshop presentation, 40% final exam + 50% project + 10% report/meetings)Announcements
Suggested Reading
Main textbook:
Networks of the Brain by Olaf Sporns, The MIT Press ISBN 978-0-262-01469-4
Supplementary textbooks:
Νευροεπιστήμη και συμπεριφορά. Eric Kandel, James Schwartz, and Thomas Jessell (Μετάφραση στα ελληνικά, Χάρης Καζλαρής και άλλοι, Πανεπιστημιακές Εκδόσεις Κρήτης)
Βασικές αρχές λειτουργίας του νευρικού συστήματος. Kiki Sidiropoulou
References - Papers (to be extended)
Spontaneous behaviors drive multidimensional, brainwide activity Carsen Stringer, Marius Pachitariu, Nicholas Steinmetz, Charu Bai Reddy, Matteo Carandini, Kenneth D. Harris
Evolving the Olfactory System Guangyu Robert Yang, Peter Yiliu Wang, Yi Sun, Ashok Litwin-Kumar, Richard Axel, L.F. Abbott
Endogenous Sequential Cortical Activity Evoked by Visual Stimuli Luis Carrillo-Reid, Jae-eun Kang Miller, Jordan P. Hamm, Jesse Jackson, and Rafael Yuste
Imprinting and recalling cortical ensembles Luis Carrillo-Reid, Weijian Yang, Yuki Bando, Darcy S. Peterka, Rafael Yuste
Cellular imaging of visual cortex reveals the spatial and functional organization of spontaneous activity Yeang H. Chng and R. Clay Reid
Spontaneous Events Outline the Realm of Possible Sensory Responses in Neocortical Populations Artur Luczak, Peter Bartho and Kenneth D. Harris
Visual stimuli recruit intrinsically generated cortical ensembles Jae-eun Kang Miller, Inbal Ayzenshtat, Luis Carrillo-Reid, and Rafael Yuste
Internally Mediated Developmental Desynchronization of Neocortical Network Activty. Golshani P., Goncalves J.T. , Khoshkhoo S., Mostany R, Smirnakis S. and Portera-Cailliau C.
Attractor dynamics of network UP states in the neocortex. Cossart R., Aronov D., and Yuste R.
Detecting pairwise correlations in spike trains: an objective comparison of methods and application to the study of retinal waves. Cutts, C.S. and S.J. Eglen. J Neurosci, 2014. 34(43): p. 14288-303.
Anatomy and function of an excitatory network in the visual cortex. R. Clay Reed. Nature 532: 370-374 (2016).
Functional organization of excitatory synaptic strength in primary visualcortex. Cossell, L., et al. Nature 518.7539 (2015): 399.
Nature. 2015 May 28;521(7553):436-44. doi: 10.1038/nature14539
A functional microcircuit for cat visual cortex. Douglas RJ, Martin KA. J Physiol. 1991;440:735-69.
Opening the grey box. Douglas RJ, Martin KA.Trends Neurosci. 1991 Jul;14(7):286-93. Review.PMID: 1719675
Mapping the matrix: the ways of neocortex. Douglas RJ, Martin KA.Neuron. 2007 Oct 25;56(2):226-38. Review. PMID: 17964242
Topology and dynamics of the canonical circuit of cat V1. Binzegger T, Douglas RJ, Martin KA. Neural Netw. 2009
Class-specific features of neuronal wiring. Stepanyants A, Tamas G, Chklovskii DB.Neuron. 2004 Jul 22;43(2):251-9.PMID: 15260960
Principles of Connectivity among Morphologically Defined Cell Types in Adult Neocortex. Science. Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S & Tolias AS (2015). Vol. 350 no. 6264 DOI: 10.1126 Link
Small World/Scale free:Small-world Propensity and Weighted Brain Networks. Muldoon S., Bridgeford E.W., Bassett D.S.
Collective dynamics of 'small-world' networks. Watts D. J., Strogatz S. H.
Network 'Small-World-Ness': A Quantitative Method for Determining Canonical Network Equivalence. Humphries M.D., Gurney K.
Scale-free characteristics of random networks: the topology of the world-wide web. Barabasi A., Albert R., Jeong H.
Networks Science, The Scale-free Property (section 4), Ablert-Laszlo Barabasi
Collective dynamics of 'small-world' networks. Watts DJ, Strogatz SH. Nature 1998;393(6684):440-2.
Deep Learning:Deep learning. LeCun Y1, Bengio Y2, Hinton G3.
Toward an Integration of Deep Learning and Neuroscience. Adam H. Marblestone1, Greg Wayne and Konrad P. Kording
Methods of Deconvolution:[Note: Historically Friedrich came first, then Vogelstein. Essentially simultaneously with Sang, Pnevmatikakis proposed another method which is now the most currently used method, and which Anna Palagina employed for the datasets of the projects.]
Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Yaksi E., Friedrich R.W.
Fast Nonnegative Deconvolution for Spike Train Inference From Population Calcium Imaging. Vogelstein J.T., Packer A.M., Machado T.A., et al.
Visually Driven Neuropil Activity and Information Encoding in Mouse Primary Visual Cortex. Sangkyun L., Meyer J., Park J., Smirnakis S.M.
Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Pnevmatikakis E.A., Soudry D., Gao Y., et al.
Preparation for the final Exam:
Neuroscience Basics: Neuroscience Basics - Questions.Datasets, Videos and Other Resources
BrainMap BrainMap database of published functional and structural neuroimaging experiments with coordinate-based results (x,y,z) in Talairach or MNI space. The goal of BrainMap is to develop software and tools to share neuroimaging results and enable meta-analysis of studies of human brain function and structure in healthy and diseased subjects. http://brainmap.org
Allen Brain Map Accelerating progress toward understanding the brain. https://portal.brain-map.org/
Human Brain Project The Human Brain Project aims to put in place a cutting-edge research infrastructure that will allow scientific and industrial researchers to advance our knowledge in the fields of neuroscience, computing, and brain-related medicine. https://www.humanbrainproject.eu
Brain Drop A Mixed-Signal Neuromorphic Architecture With a Dynamical Systems-Based Programming Model. https://ieeexplore.ieee.org/document/8591981
Tentative Assignments
| Week | Short description |
| 1-10 | Reading papers/textbook |
| 3 | Graph theoretical metrics, simple statistics (CDFs, histograms) |
| 4 | Reading papers |
| 5 | Null hypothesis, temporal correlation |
| 6 | Project |
| 6-9 | Group meetings - milestones |
| 10 | Presentations |
|
Projects
Identification and start of the implementation of the project from 1) literature review, or 2) a small data analysis laboratory project on two photon data that will be provided. The project could involve the graph-theoretical analysis and pattern discovery of neuronal circuits in absence epilepsy, under spontaneous conditions, etc Team work under the supervision of Stelios Smirnakis & Maria Papadopouli |
Marking scheme
| Midterm | N/A |
| Assignments | ✔ |
| Final exam | ✔ |
| Project | Customized to each team |
Other Sources:
A journey through the Visual System
Exploring the Visual Brain
From Single Cells to Whole Brains: Gene Expression, Cell Types, and the Brain:

